Outliers are imputed with some representative values and statistical methods.
imputate_outlier(.data, xvar, method, no_attrs, cap_ntiles)Arguments
- .data
a data.frame or a
tbl_df.- xvar
variable name to replace missing value.
- method
method of missing values imputation.
- no_attrs
logical. If TRUE, return numerical variable or categorical variable. else If FALSE, imputation class.
- cap_ntiles
numeric. Only used when method is "capping". Specifies the value of percentiles replaced by the values of lower outliers and upper outliers. The default is c(0.05, 0.95).
Value
An object of imputation class. or numerical variable. if no_attrs is FALSE then return imputation class, else no_attrs is TRUE then return numerical vector. Attributes of imputation class is as follows.
method : method of missing value imputation.
predictor is numerical variable
"mean" : arithmetic mean
"median" : median
"mode" : mode
"capping" : Impute the upper outliers with 95 percentile, and Impute the lower outliers with 5 percentile.
You can change this criterion with the cap_ntiles argument.
outlier_pos : position of outliers in predictor.
outliers : outliers. outliers corresponding to outlier_pos.
type : "outliers". type of imputation.
Details
imputate_outlier() creates an imputation class. The `imputation` class includes missing value position, imputed value, and method of missing value imputation, etc. The `imputation` class compares the imputed value with the original value to help determine whether the imputed value is used in the analysis.
See vignette("transformation") for an introduction to these concepts.
See also
Examples
# \donttest{
# Replace the outliers of the sodium variable with median.
imputate_outlier(heartfailure, sodium, method = "median")
#> [1] 130 136 129 137 137 132 137 131 138 133 131 140 137 137 138 136 140 127
#> [19] 140 137 137 136 140 135 134 144 138 136 134 132 134 132 128 138 140 134
#> [37] 134 145 137 142 134 136 139 134 142 135 130 138 133 140 138 139 146 134
#> [55] 132 132 138 138 136 136 139 131 139 145 137 127 136 140 142 135 140 139
#> [73] 132 137 134 139 140 140 131 140 136 137 132 133 141 140 137 140 139 144
#> [91] 136 133 137 135 142 141 134 136 137 140 141 137 144 140 143 138 137 138
#> [109] 133 142 132 135 136 137 126 139 136 138 140 134 135 136 140 145 134 135
#> [127] 137 137 136 145 138 131 137 145 137 137 137 130 136 138 134 140 132 141
#> [145] 139 141 136 137 134 136 135 139 134 137 136 140 136 136 134 139 134 139
#> [163] 137 142 139 135 133 134 138 133 136 140 145 139 137 138 135 140 145 140
#> [181] 136 136 136 134 137 136 134 144 136 140 134 135 130 142 135 145 137 138
#> [199] 134 137 137 136 138 136 137 136 141 136 141 130 136 140 138 130 134 134
#> [217] 141 137 134 140 134 145 139 134 138 125 132 140 130 134 127 134 137 137
#> [235] 137 137 148 132 136 137 139 136 136 141 134 137 138 135 142 130 139 143
#> [253] 133 132 139 142 139 139 135 135 138 133 129 140 141 140 134 140 140 132
#> [271] 130 134 133 140 137 137 142 140 136 136 137 136 128 138 141 135 140 132
#> [289] 142 144 141 139 140 136 143 139 138 140 136
#> attr(,"method")
#> [1] "median"
#> attr(,"var_type")
#> [1] "numerical"
#> attr(,"outlier_pos")
#> [1] 5 20 127 200
#> attr(,"outliers")
#> [1] 116 121 124 113
#> attr(,"type")
#> [1] "outliers"
#> attr(,"message")
#> [1] "complete imputation"
#> attr(,"success")
#> [1] TRUE
#> attr(,"class")
#> [1] "imputation" "numeric"
# Replace the outliers of the sodium variable with capping.
imputate_outlier(heartfailure, sodium, method = "capping")
#> [1] 130 136 129 137 130 132 137 131 138 133 131 140 137 137 138 136 140 127
#> [19] 140 130 137 136 140 135 134 144 138 136 134 132 134 132 128 138 140 134
#> [37] 134 145 137 142 134 136 139 134 142 135 130 138 133 140 138 139 146 134
#> [55] 132 132 138 138 136 136 139 131 139 145 137 127 136 140 142 135 140 139
#> [73] 132 137 134 139 140 140 131 140 136 137 132 133 141 140 137 140 139 144
#> [91] 136 133 137 135 142 141 134 136 137 140 141 137 144 140 143 138 137 138
#> [109] 133 142 132 135 136 137 126 139 136 138 140 134 135 136 140 145 134 135
#> [127] 130 137 136 145 138 131 137 145 137 137 137 130 136 138 134 140 132 141
#> [145] 139 141 136 137 134 136 135 139 134 137 136 140 136 136 134 139 134 139
#> [163] 137 142 139 135 133 134 138 133 136 140 145 139 137 138 135 140 145 140
#> [181] 136 136 136 134 137 136 134 144 136 140 134 135 130 142 135 145 137 138
#> [199] 134 130 137 136 138 136 137 136 141 136 141 130 136 140 138 130 134 134
#> [217] 141 137 134 140 134 145 139 134 138 125 132 140 130 134 127 134 137 137
#> [235] 137 137 148 132 136 137 139 136 136 141 134 137 138 135 142 130 139 143
#> [253] 133 132 139 142 139 139 135 135 138 133 129 140 141 140 134 140 140 132
#> [271] 130 134 133 140 137 137 142 140 136 136 137 136 128 138 141 135 140 132
#> [289] 142 144 141 139 140 136 143 139 138 140 136
#> attr(,"method")
#> [1] "capping"
#> attr(,"var_type")
#> [1] "numerical"
#> attr(,"outlier_pos")
#> [1] 5 20 127 200
#> attr(,"outliers")
#> [1] 116 121 124 113
#> attr(,"type")
#> [1] "outliers"
#> attr(,"message")
#> [1] "complete imputation"
#> attr(,"success")
#> [1] TRUE
#> attr(,"class")
#> [1] "imputation" "numeric"
imputate_outlier(heartfailure, sodium, method = "capping",
cap_ntiles = c(0.1, 0.9))
#> [1] 130 136 129 137 132 132 137 131 138 133 131 140 137 137 138 136 140 127
#> [19] 140 132 137 136 140 135 134 144 138 136 134 132 134 132 128 138 140 134
#> [37] 134 145 137 142 134 136 139 134 142 135 130 138 133 140 138 139 146 134
#> [55] 132 132 138 138 136 136 139 131 139 145 137 127 136 140 142 135 140 139
#> [73] 132 137 134 139 140 140 131 140 136 137 132 133 141 140 137 140 139 144
#> [91] 136 133 137 135 142 141 134 136 137 140 141 137 144 140 143 138 137 138
#> [109] 133 142 132 135 136 137 126 139 136 138 140 134 135 136 140 145 134 135
#> [127] 132 137 136 145 138 131 137 145 137 137 137 130 136 138 134 140 132 141
#> [145] 139 141 136 137 134 136 135 139 134 137 136 140 136 136 134 139 134 139
#> [163] 137 142 139 135 133 134 138 133 136 140 145 139 137 138 135 140 145 140
#> [181] 136 136 136 134 137 136 134 144 136 140 134 135 130 142 135 145 137 138
#> [199] 134 132 137 136 138 136 137 136 141 136 141 130 136 140 138 130 134 134
#> [217] 141 137 134 140 134 145 139 134 138 125 132 140 130 134 127 134 137 137
#> [235] 137 137 148 132 136 137 139 136 136 141 134 137 138 135 142 130 139 143
#> [253] 133 132 139 142 139 139 135 135 138 133 129 140 141 140 134 140 140 132
#> [271] 130 134 133 140 137 137 142 140 136 136 137 136 128 138 141 135 140 132
#> [289] 142 144 141 139 140 136 143 139 138 140 136
#> attr(,"method")
#> [1] "capping"
#> attr(,"var_type")
#> [1] "numerical"
#> attr(,"outlier_pos")
#> [1] 5 20 127 200
#> attr(,"outliers")
#> [1] 116 121 124 113
#> attr(,"type")
#> [1] "outliers"
#> attr(,"message")
#> [1] "complete imputation"
#> attr(,"success")
#> [1] TRUE
#> attr(,"class")
#> [1] "imputation" "numeric"
## using dplyr -------------------------------------
library(dplyr)
# The mean before and after the imputation of the sodium variable
heartfailure %>%
mutate(sodium_imp = imputate_outlier(heartfailure, sodium,
method = "capping", no_attrs = TRUE)) %>%
group_by(death_event) %>%
summarise(orig = mean(sodium, na.rm = TRUE),
imputation = mean(sodium_imp, na.rm = TRUE))
#> # A tibble: 2 × 3
#> death_event orig imputation
#> <fct> <dbl> <dbl>
#> 1 No 137. 137.
#> 2 Yes 135. 136.
# If the variable of interest is a numerical variables
sodium <- imputate_outlier(heartfailure, sodium)
sodium
#> [1] 130 136 129 137 130 132 137 131 138 133 131 140 137 137 138 136 140 127
#> [19] 140 130 137 136 140 135 134 144 138 136 134 132 134 132 128 138 140 134
#> [37] 134 145 137 142 134 136 139 134 142 135 130 138 133 140 138 139 146 134
#> [55] 132 132 138 138 136 136 139 131 139 145 137 127 136 140 142 135 140 139
#> [73] 132 137 134 139 140 140 131 140 136 137 132 133 141 140 137 140 139 144
#> [91] 136 133 137 135 142 141 134 136 137 140 141 137 144 140 143 138 137 138
#> [109] 133 142 132 135 136 137 126 139 136 138 140 134 135 136 140 145 134 135
#> [127] 130 137 136 145 138 131 137 145 137 137 137 130 136 138 134 140 132 141
#> [145] 139 141 136 137 134 136 135 139 134 137 136 140 136 136 134 139 134 139
#> [163] 137 142 139 135 133 134 138 133 136 140 145 139 137 138 135 140 145 140
#> [181] 136 136 136 134 137 136 134 144 136 140 134 135 130 142 135 145 137 138
#> [199] 134 130 137 136 138 136 137 136 141 136 141 130 136 140 138 130 134 134
#> [217] 141 137 134 140 134 145 139 134 138 125 132 140 130 134 127 134 137 137
#> [235] 137 137 148 132 136 137 139 136 136 141 134 137 138 135 142 130 139 143
#> [253] 133 132 139 142 139 139 135 135 138 133 129 140 141 140 134 140 140 132
#> [271] 130 134 133 140 137 137 142 140 136 136 137 136 128 138 141 135 140 132
#> [289] 142 144 141 139 140 136 143 139 138 140 136
#> attr(,"method")
#> [1] "capping"
#> attr(,"var_type")
#> [1] "numerical"
#> attr(,"outlier_pos")
#> [1] 5 20 127 200
#> attr(,"outliers")
#> [1] 116 121 124 113
#> attr(,"type")
#> [1] "outliers"
#> attr(,"message")
#> [1] "complete imputation"
#> attr(,"success")
#> [1] TRUE
#> attr(,"class")
#> [1] "imputation" "numeric"
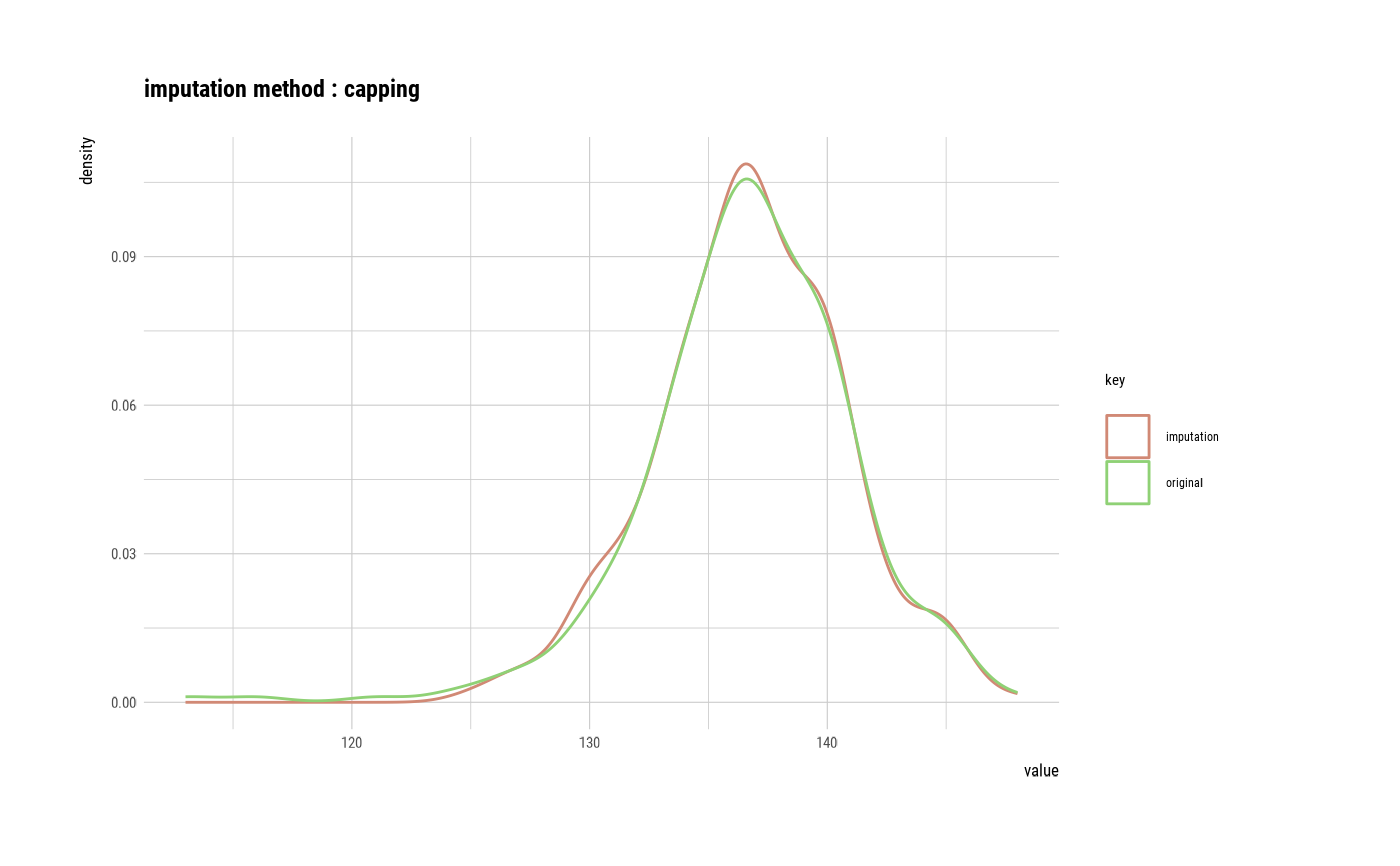
summary(sodium)
#> Impute outliers with capping
#>
#> * Information of Imputation (before vs after)
#> Original Imputation
#> described_variables "value" "value"
#> n "299" "299"
#> na "0" "0"
#> mean "136.6254" "136.7793"
#> sd "4.412477" "3.921816"
#> se_mean "0.2551802" "0.2268045"
#> IQR "6" "6"
#> skewness "-1.0481360" "-0.1201672"
#> kurtosis "4.119712" "0.189418"
#> p00 "113" "125"
#> p01 "123.94" "127.00"
#> p05 "130" "130"
#> p10 "132" "132"
#> p20 "134" "134"
#> p25 "134" "134"
#> p30 "135" "135"
#> p40 "136" "136"
#> p50 "137" "137"
#> p60 "138" "138"
#> p70 "139" "139"
#> p75 "140" "140"
#> p80 "140" "140"
#> p90 "141.2" "141.2"
#> p95 "144" "144"
#> p99 "145" "145"
#> p100 "148" "148"
plot(sodium)
 # }
# }