Visualize two kinds of plot by attribute of `transform` class. The transformation of a numerical variable is a density plot.
# S3 method for class 'transform'
plot(x, typographic = TRUE, base_family = NULL, ...)Arguments
- x
an object of class "transform", usually, a result of a call to transform().
- typographic
logical. Whether to apply focuses on typographic elements to ggplot2 visualization. The default is TRUE. if TRUE provides a base theme that focuses on typographic elements.
- base_family
character. The name of the base font family to use for the visualization. If not specified, the font defined in dlookr is applied. (See details)
- ...
arguments to be passed to methods, such as graphical parameters (see par).
Details
The base_family is selected from "Roboto Condensed", "Liberation Sans Narrow", "NanumSquare", "Noto Sans Korean". If you want to use a different font, use it after loading the Google font with import_google_font().
See also
Examples
# \donttest{
# Standardization ------------------------------
creatinine_minmax <- transform(heartfailure$creatinine, method = "minmax")
creatinine_minmax
#> [1] 0.15730337 0.06741573 0.08988764 0.15730337 0.24719101 0.17977528
#> [7] 0.07865169 0.06741573 0.11235955 1.00000000 0.39325843 0.04494382
#> [13] 0.06741573 0.06741573 0.05617978 0.08988764 0.04494382 0.03370787
#> [19] 0.05617978 0.15730337 0.08988764 0.12359551 0.04494382 0.03370787
#> [25] 0.14943820 0.15730337 0.05617978 0.08988764 0.59550562 0.07865169
#> [31] 0.14943820 0.28089888 0.05617978 0.07865169 0.05617978 0.33707865
#> [37] 0.05617978 0.05617978 0.20224719 0.28089888 0.14943820 0.07865169
#> [43] 0.07865169 0.05617978 0.06741573 0.15730337 0.04494382 0.01123596
#> [49] 0.43820225 0.05617978 0.05617978 0.10112360 0.70786517 0.05617978
#> [55] 0.19101124 0.16853933 0.24719101 0.01123596 0.06741573 0.08988764
#> [61] 0.05617978 0.20224719 0.06741573 0.05617978 0.07640449 0.26966292
#> [67] 0.08988764 0.05617978 0.07865169 0.14943820 0.03370787 0.04494382
#> [73] 0.05617978 0.08988764 0.07865169 0.02247191 0.03370787 0.07865169
#> [79] 0.01123596 0.04494382 0.13483146 0.07640449 0.22471910 0.14606742
#> [85] 0.05617978 0.02247191 0.06741573 0.03370787 0.02247191 0.06741573
#> [91] 0.03370787 0.05617978 0.07640449 0.13483146 0.02247191 0.05617978
#> [97] 0.08988764 0.06741573 0.07865169 0.06741573 0.06741573 0.07640449
#> [103] 0.06741573 0.05617978 0.20224719 0.13483146 0.08988764 0.04494382
#> [109] 0.06741573 0.08988764 0.07865169 0.07865169 0.12359551 0.08988764
#> [115] 0.07865169 0.05617978 0.02247191 0.30337079 0.04494382 0.14943820
#> [121] 0.11235955 0.05617978 0.02808989 0.04494382 0.35955056 0.08988764
#> [127] 0.17977528 0.03370787 0.02247191 0.32584270 0.02247191 0.62921348
#> [133] 0.07640449 0.08988764 0.07640449 0.07640449 0.04494382 0.17977528
#> [139] 0.05617978 0.03370787 0.06741573 0.04494382 0.04494382 0.04494382
#> [145] 0.13483146 0.02247191 0.02247191 0.05617978 0.14943820 0.04494382
#> [151] 0.22471910 0.04494382 0.04494382 0.07640449 0.03370787 0.13483146
#> [157] 0.10112360 0.05617978 0.08988764 0.06741573 0.07865169 0.03370787
#> [163] 0.04494382 0.04494382 0.06741573 0.08988764 0.02247191 0.21348315
#> [169] 0.05617978 0.03370787 0.11235955 0.04494382 0.06741573 0.03370787
#> [175] 0.04494382 0.05617978 0.05617978 0.05617978 0.07865169 0.02247191
#> [181] 0.04494382 0.05617978 0.07865169 0.22471910 0.07865169 0.11235955
#> [187] 0.01123596 0.17977528 0.05617978 0.04494382 0.17977528 0.11235955
#> [193] 0.02247191 0.07640449 0.12359551 0.14606742 0.07640449 0.03370787
#> [199] 0.05617978 0.14606742 0.02247191 0.05617978 0.04494382 0.33707865
#> [205] 0.02247191 0.05617978 0.03370787 0.04494382 0.05617978 0.03370787
#> [211] 0.05617978 0.03370787 0.10112360 0.12359551 0.03370787 0.08988764
#> [217] 0.04494382 0.95505618 0.06741573 0.02247191 0.14943820 0.06741573
#> [223] 0.06741573 0.03370787 0.05617978 0.10112360 0.08988764 0.05617978
#> [229] 0.50561798 0.07865169 0.13483146 0.06741573 0.04494382 0.10112360
#> [235] 0.06741573 0.06741573 0.06741573 0.07865169 0.05617978 0.07640449
#> [241] 0.08988764 0.08988764 0.06741573 0.04494382 0.14606742 0.10112360
#> [247] 0.06741573 0.21348315 0.05617978 0.07865169 0.00000000 0.03370787
#> [253] 0.05617978 0.07865169 0.05617978 0.05617978 0.13483146 0.05617978
#> [259] 0.03370787 0.02247191 0.05617978 0.02247191 0.10112360 0.05617978
#> [265] 0.07865169 0.04494382 0.14943820 0.13483146 0.04494382 0.05617978
#> [271] 0.12359551 0.04494382 0.07865169 0.02247191 0.05617978 0.03370787
#> [277] 0.06741573 0.06741573 0.02247191 0.08988764 0.05617978 0.24719101
#> [283] 0.37078652 0.06741573 0.03370787 0.07865169 0.13483146 0.05617978
#> [289] 0.06741573 0.04494382 0.03370787 0.10112360 0.05617978 0.04494382
#> [295] 0.06741573 0.07865169 0.03370787 0.10112360 0.12359551
summary(creatinine_minmax)
#> * Standardization with minmax
#>
#> * Information of Transformation (before vs after)
#> Original Transformation
#> n 299.00000000 2.990000e+02
#> na 0.00000000 0.000000e+00
#> mean 1.39387960 1.004359e-01
#> sd 1.03451006 1.162371e-01
#> se_mean 0.05982726 6.722164e-03
#> IQR 0.50000000 5.617978e-02
#> skewness 4.45599588 4.455996e+00
#> kurtosis 25.82823866 2.582824e+01
#> p00 0.50000000 0.000000e+00
#> p01 0.60000000 1.123596e-02
#> p05 0.70000000 2.247191e-02
#> p10 0.80000000 3.370787e-02
#> p20 0.90000000 4.494382e-02
#> p25 0.90000000 4.494382e-02
#> p30 1.00000000 5.617978e-02
#> p40 1.00000000 5.617978e-02
#> p50 1.10000000 6.741573e-02
#> p60 1.20000000 7.865169e-02
#> p70 1.30000000 8.988764e-02
#> p75 1.40000000 1.011236e-01
#> p80 1.70000000 1.348315e-01
#> p90 2.10000000 1.797753e-01
#> p95 3.00000000 2.808989e-01
#> p99 6.11400000 6.307865e-01
#> p100 9.40000000 1.000000e+00
plot(creatinine_minmax)
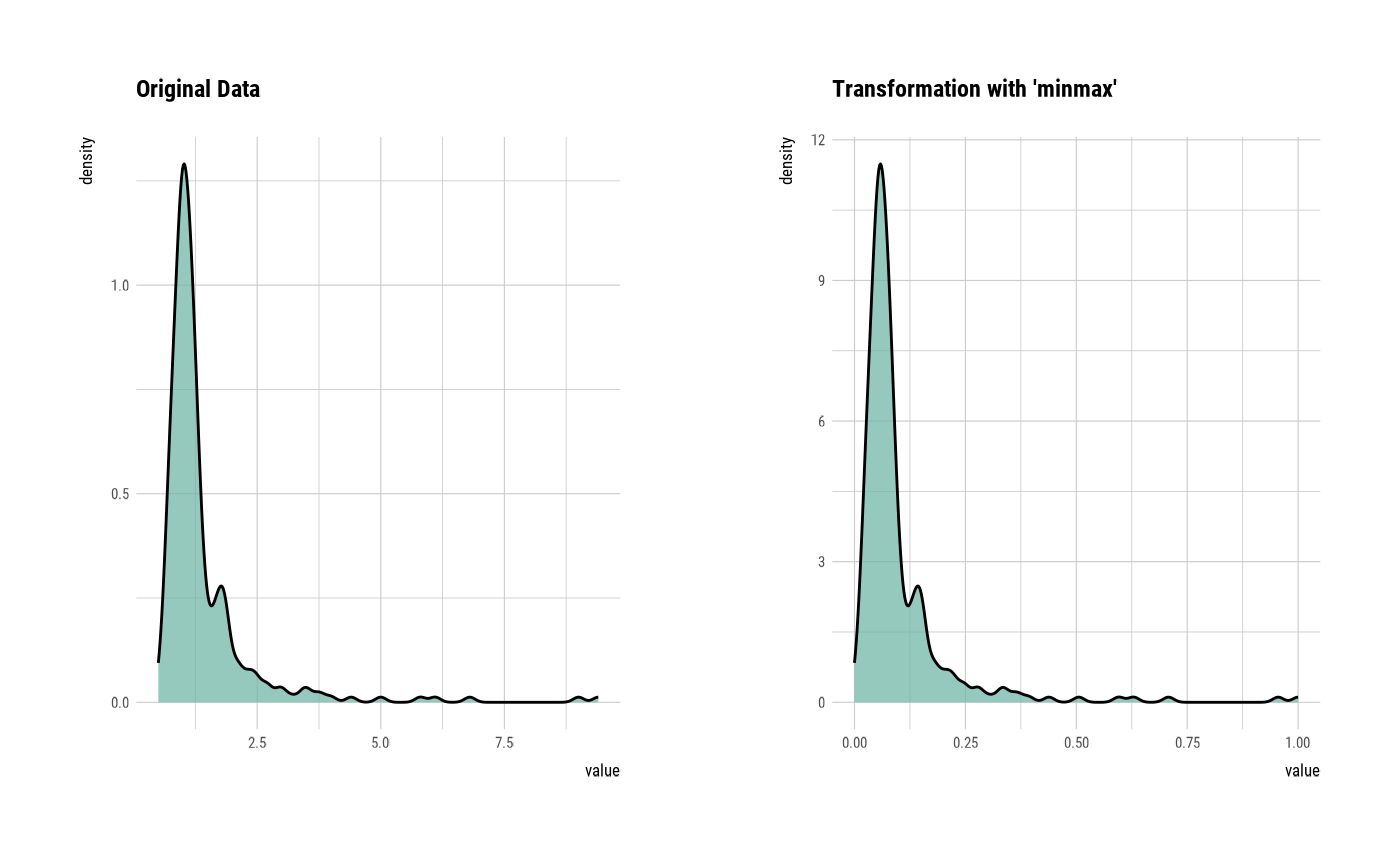 # Resolving Skewness --------------------------
creatinine_log <- transform(heartfailure$creatinine, method = "log")
creatinine_log
#> [1] 0.64185389 0.09531018 0.26236426 0.64185389 0.99325177 0.74193734
#> [7] 0.18232156 0.09531018 0.40546511 2.24070969 1.38629436 -0.10536052
#> [13] 0.09531018 0.09531018 0.00000000 0.26236426 -0.10536052 -0.22314355
#> [19] 0.00000000 0.64185389 0.26236426 0.47000363 -0.10536052 -0.22314355
#> [25] 0.60431597 0.64185389 0.00000000 0.26236426 1.75785792 0.18232156
#> [31] 0.60431597 1.09861229 0.00000000 0.18232156 0.00000000 1.25276297
#> [37] 0.00000000 0.00000000 0.83290912 1.09861229 0.60431597 0.18232156
#> [43] 0.18232156 0.00000000 0.09531018 0.64185389 -0.10536052 -0.51082562
#> [49] 1.48160454 0.00000000 0.00000000 0.33647224 1.91692261 0.00000000
#> [55] 0.78845736 0.69314718 0.99325177 -0.51082562 0.09531018 0.26236426
#> [61] 0.00000000 0.83290912 0.09531018 0.00000000 0.16551444 1.06471074
#> [67] 0.26236426 0.00000000 0.18232156 0.60431597 -0.22314355 -0.10536052
#> [73] 0.00000000 0.26236426 0.18232156 -0.35667494 -0.22314355 0.18232156
#> [79] -0.51082562 -0.10536052 0.53062825 0.16551444 0.91629073 0.58778666
#> [85] 0.00000000 -0.35667494 0.09531018 -0.22314355 -0.35667494 0.09531018
#> [91] -0.22314355 0.00000000 0.16551444 0.53062825 -0.35667494 0.00000000
#> [97] 0.26236426 0.09531018 0.18232156 0.09531018 0.09531018 0.16551444
#> [103] 0.09531018 0.00000000 0.83290912 0.53062825 0.26236426 -0.10536052
#> [109] 0.09531018 0.26236426 0.18232156 0.18232156 0.47000363 0.26236426
#> [115] 0.18232156 0.00000000 -0.35667494 1.16315081 -0.10536052 0.60431597
#> [121] 0.40546511 0.00000000 -0.28768207 -0.10536052 1.30833282 0.26236426
#> [127] 0.74193734 -0.22314355 -0.35667494 1.22377543 -0.35667494 1.80828877
#> [133] 0.16551444 0.26236426 0.16551444 0.16551444 -0.10536052 0.74193734
#> [139] 0.00000000 -0.22314355 0.09531018 -0.10536052 -0.10536052 -0.10536052
#> [145] 0.53062825 -0.35667494 -0.35667494 0.00000000 0.60431597 -0.10536052
#> [151] 0.91629073 -0.10536052 -0.10536052 0.16551444 -0.22314355 0.53062825
#> [157] 0.33647224 0.00000000 0.26236426 0.09531018 0.18232156 -0.22314355
#> [163] -0.10536052 -0.10536052 0.09531018 0.26236426 -0.35667494 0.87546874
#> [169] 0.00000000 -0.22314355 0.40546511 -0.10536052 0.09531018 -0.22314355
#> [175] -0.10536052 0.00000000 0.00000000 0.00000000 0.18232156 -0.35667494
#> [181] -0.10536052 0.00000000 0.18232156 0.91629073 0.18232156 0.40546511
#> [187] -0.51082562 0.74193734 0.00000000 -0.10536052 0.74193734 0.40546511
#> [193] -0.35667494 0.16551444 0.47000363 0.58778666 0.16551444 -0.22314355
#> [199] 0.00000000 0.58778666 -0.35667494 0.00000000 -0.10536052 1.25276297
#> [205] -0.35667494 0.00000000 -0.22314355 -0.10536052 0.00000000 -0.22314355
#> [211] 0.00000000 -0.22314355 0.33647224 0.47000363 -0.22314355 0.26236426
#> [217] -0.10536052 2.19722458 0.09531018 -0.35667494 0.60431597 0.09531018
#> [223] 0.09531018 -0.22314355 0.00000000 0.33647224 0.26236426 0.00000000
#> [229] 1.60943791 0.18232156 0.53062825 0.09531018 -0.10536052 0.33647224
#> [235] 0.09531018 0.09531018 0.09531018 0.18232156 0.00000000 0.16551444
#> [241] 0.26236426 0.26236426 0.09531018 -0.10536052 0.58778666 0.33647224
#> [247] 0.09531018 0.87546874 0.00000000 0.18232156 -0.69314718 -0.22314355
#> [253] 0.00000000 0.18232156 0.00000000 0.00000000 0.53062825 0.00000000
#> [259] -0.22314355 -0.35667494 0.00000000 -0.35667494 0.33647224 0.00000000
#> [265] 0.18232156 -0.10536052 0.60431597 0.53062825 -0.10536052 0.00000000
#> [271] 0.47000363 -0.10536052 0.18232156 -0.35667494 0.00000000 -0.22314355
#> [277] 0.09531018 0.09531018 -0.35667494 0.26236426 0.00000000 0.99325177
#> [283] 1.33500107 0.09531018 -0.22314355 0.18232156 0.53062825 0.00000000
#> [289] 0.09531018 -0.10536052 -0.22314355 0.33647224 0.00000000 -0.10536052
#> [295] 0.09531018 0.18232156 -0.22314355 0.33647224 0.47000363
summary(creatinine_log)
#> * Resolving Skewness with log
#>
#> * Information of Transformation (before vs after)
#> Original Transformation
#> n 299.00000000 299.00000000
#> na 0.00000000 0.00000000
#> mean 1.39387960 0.19858693
#> sd 1.03451006 0.45310789
#> se_mean 0.05982726 0.02620391
#> IQR 0.50000000 0.44183275
#> skewness 4.45599588 1.58398978
#> kurtosis 25.82823866 3.60164397
#> p00 0.50000000 -0.69314718
#> p01 0.60000000 -0.51082562
#> p05 0.70000000 -0.35667494
#> p10 0.80000000 -0.22314355
#> p20 0.90000000 -0.10536052
#> p25 0.90000000 -0.10536052
#> p30 1.00000000 0.00000000
#> p40 1.00000000 0.00000000
#> p50 1.10000000 0.09531018
#> p60 1.20000000 0.18232156
#> p70 1.30000000 0.26236426
#> p75 1.40000000 0.33647224
#> p80 1.70000000 0.53062825
#> p90 2.10000000 0.74193734
#> p95 3.00000000 1.09861229
#> p99 6.11400000 1.81046145
#> p100 9.40000000 2.24070969
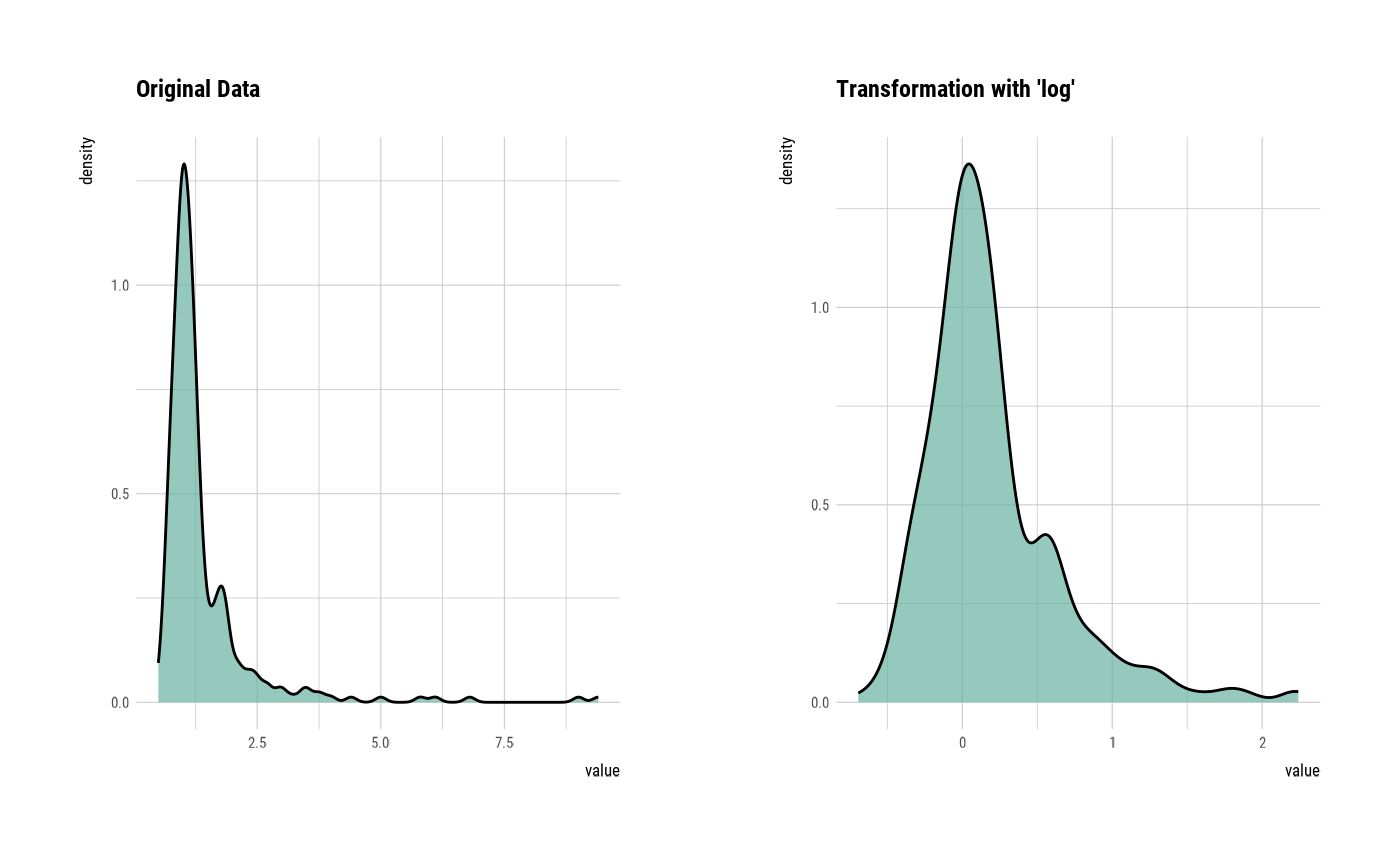
plot(creatinine_log)
# Resolving Skewness --------------------------
creatinine_log <- transform(heartfailure$creatinine, method = "log")
creatinine_log
#> [1] 0.64185389 0.09531018 0.26236426 0.64185389 0.99325177 0.74193734
#> [7] 0.18232156 0.09531018 0.40546511 2.24070969 1.38629436 -0.10536052
#> [13] 0.09531018 0.09531018 0.00000000 0.26236426 -0.10536052 -0.22314355
#> [19] 0.00000000 0.64185389 0.26236426 0.47000363 -0.10536052 -0.22314355
#> [25] 0.60431597 0.64185389 0.00000000 0.26236426 1.75785792 0.18232156
#> [31] 0.60431597 1.09861229 0.00000000 0.18232156 0.00000000 1.25276297
#> [37] 0.00000000 0.00000000 0.83290912 1.09861229 0.60431597 0.18232156
#> [43] 0.18232156 0.00000000 0.09531018 0.64185389 -0.10536052 -0.51082562
#> [49] 1.48160454 0.00000000 0.00000000 0.33647224 1.91692261 0.00000000
#> [55] 0.78845736 0.69314718 0.99325177 -0.51082562 0.09531018 0.26236426
#> [61] 0.00000000 0.83290912 0.09531018 0.00000000 0.16551444 1.06471074
#> [67] 0.26236426 0.00000000 0.18232156 0.60431597 -0.22314355 -0.10536052
#> [73] 0.00000000 0.26236426 0.18232156 -0.35667494 -0.22314355 0.18232156
#> [79] -0.51082562 -0.10536052 0.53062825 0.16551444 0.91629073 0.58778666
#> [85] 0.00000000 -0.35667494 0.09531018 -0.22314355 -0.35667494 0.09531018
#> [91] -0.22314355 0.00000000 0.16551444 0.53062825 -0.35667494 0.00000000
#> [97] 0.26236426 0.09531018 0.18232156 0.09531018 0.09531018 0.16551444
#> [103] 0.09531018 0.00000000 0.83290912 0.53062825 0.26236426 -0.10536052
#> [109] 0.09531018 0.26236426 0.18232156 0.18232156 0.47000363 0.26236426
#> [115] 0.18232156 0.00000000 -0.35667494 1.16315081 -0.10536052 0.60431597
#> [121] 0.40546511 0.00000000 -0.28768207 -0.10536052 1.30833282 0.26236426
#> [127] 0.74193734 -0.22314355 -0.35667494 1.22377543 -0.35667494 1.80828877
#> [133] 0.16551444 0.26236426 0.16551444 0.16551444 -0.10536052 0.74193734
#> [139] 0.00000000 -0.22314355 0.09531018 -0.10536052 -0.10536052 -0.10536052
#> [145] 0.53062825 -0.35667494 -0.35667494 0.00000000 0.60431597 -0.10536052
#> [151] 0.91629073 -0.10536052 -0.10536052 0.16551444 -0.22314355 0.53062825
#> [157] 0.33647224 0.00000000 0.26236426 0.09531018 0.18232156 -0.22314355
#> [163] -0.10536052 -0.10536052 0.09531018 0.26236426 -0.35667494 0.87546874
#> [169] 0.00000000 -0.22314355 0.40546511 -0.10536052 0.09531018 -0.22314355
#> [175] -0.10536052 0.00000000 0.00000000 0.00000000 0.18232156 -0.35667494
#> [181] -0.10536052 0.00000000 0.18232156 0.91629073 0.18232156 0.40546511
#> [187] -0.51082562 0.74193734 0.00000000 -0.10536052 0.74193734 0.40546511
#> [193] -0.35667494 0.16551444 0.47000363 0.58778666 0.16551444 -0.22314355
#> [199] 0.00000000 0.58778666 -0.35667494 0.00000000 -0.10536052 1.25276297
#> [205] -0.35667494 0.00000000 -0.22314355 -0.10536052 0.00000000 -0.22314355
#> [211] 0.00000000 -0.22314355 0.33647224 0.47000363 -0.22314355 0.26236426
#> [217] -0.10536052 2.19722458 0.09531018 -0.35667494 0.60431597 0.09531018
#> [223] 0.09531018 -0.22314355 0.00000000 0.33647224 0.26236426 0.00000000
#> [229] 1.60943791 0.18232156 0.53062825 0.09531018 -0.10536052 0.33647224
#> [235] 0.09531018 0.09531018 0.09531018 0.18232156 0.00000000 0.16551444
#> [241] 0.26236426 0.26236426 0.09531018 -0.10536052 0.58778666 0.33647224
#> [247] 0.09531018 0.87546874 0.00000000 0.18232156 -0.69314718 -0.22314355
#> [253] 0.00000000 0.18232156 0.00000000 0.00000000 0.53062825 0.00000000
#> [259] -0.22314355 -0.35667494 0.00000000 -0.35667494 0.33647224 0.00000000
#> [265] 0.18232156 -0.10536052 0.60431597 0.53062825 -0.10536052 0.00000000
#> [271] 0.47000363 -0.10536052 0.18232156 -0.35667494 0.00000000 -0.22314355
#> [277] 0.09531018 0.09531018 -0.35667494 0.26236426 0.00000000 0.99325177
#> [283] 1.33500107 0.09531018 -0.22314355 0.18232156 0.53062825 0.00000000
#> [289] 0.09531018 -0.10536052 -0.22314355 0.33647224 0.00000000 -0.10536052
#> [295] 0.09531018 0.18232156 -0.22314355 0.33647224 0.47000363
summary(creatinine_log)
#> * Resolving Skewness with log
#>
#> * Information of Transformation (before vs after)
#> Original Transformation
#> n 299.00000000 299.00000000
#> na 0.00000000 0.00000000
#> mean 1.39387960 0.19858693
#> sd 1.03451006 0.45310789
#> se_mean 0.05982726 0.02620391
#> IQR 0.50000000 0.44183275
#> skewness 4.45599588 1.58398978
#> kurtosis 25.82823866 3.60164397
#> p00 0.50000000 -0.69314718
#> p01 0.60000000 -0.51082562
#> p05 0.70000000 -0.35667494
#> p10 0.80000000 -0.22314355
#> p20 0.90000000 -0.10536052
#> p25 0.90000000 -0.10536052
#> p30 1.00000000 0.00000000
#> p40 1.00000000 0.00000000
#> p50 1.10000000 0.09531018
#> p60 1.20000000 0.18232156
#> p70 1.30000000 0.26236426
#> p75 1.40000000 0.33647224
#> p80 1.70000000 0.53062825
#> p90 2.10000000 0.74193734
#> p95 3.00000000 1.09861229
#> p99 6.11400000 1.81046145
#> p100 9.40000000 2.24070969
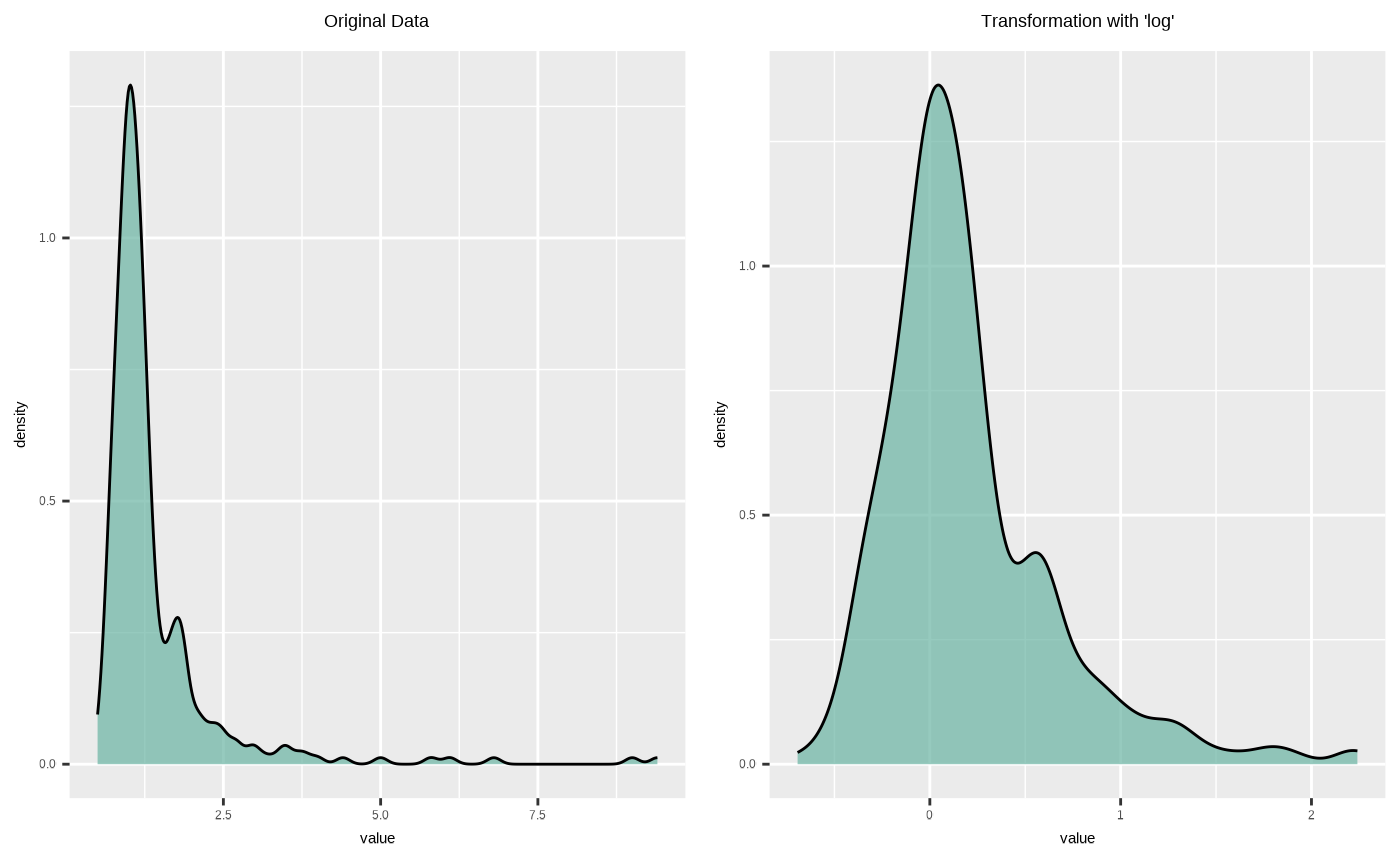
plot(creatinine_log)
 plot(creatinine_log, typographic = FALSE)
plot(creatinine_log, typographic = FALSE)
 # }
# }