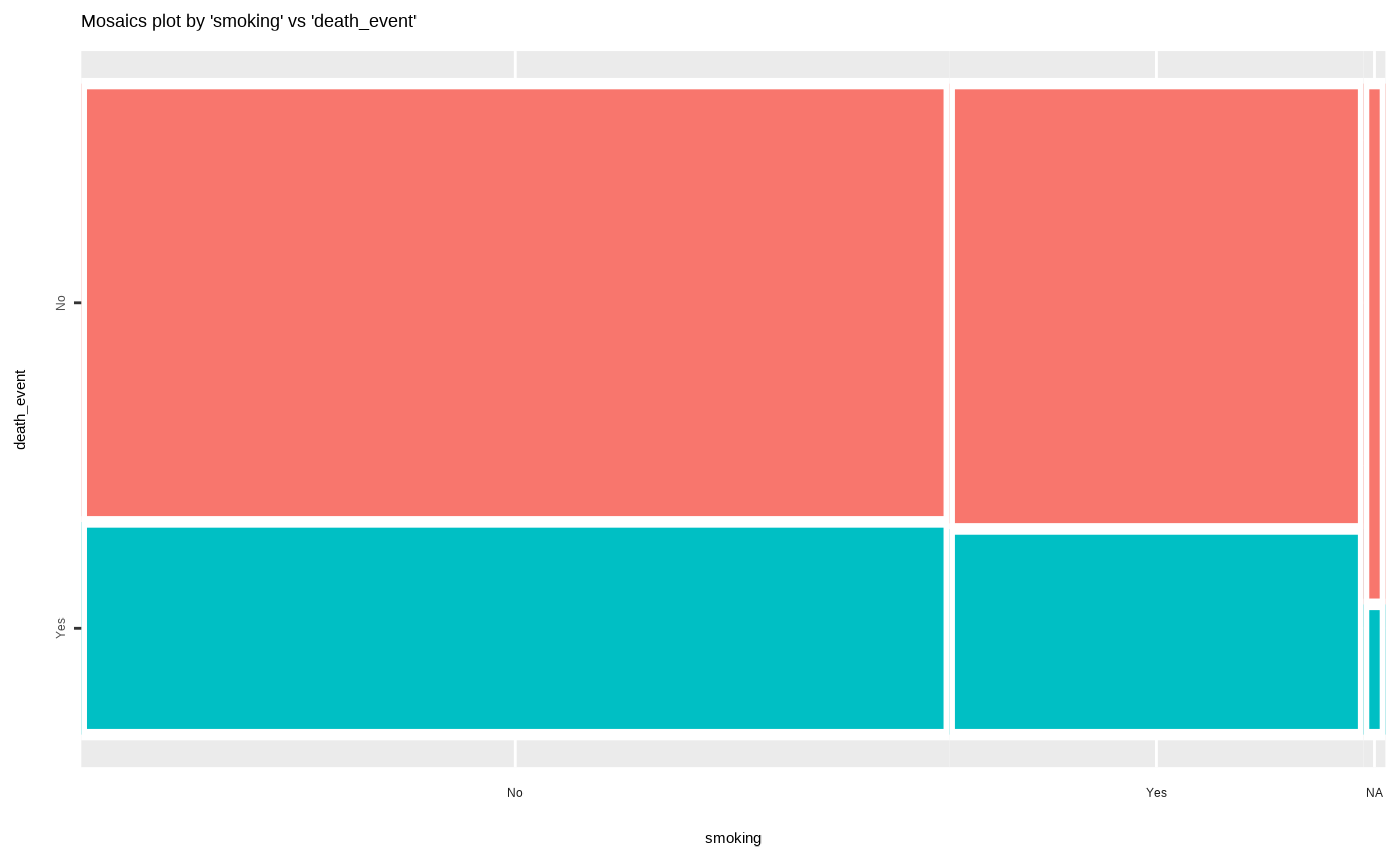
Visualize mosaics plot by attribute of compare_category class.
# S3 method for class 'compare_category'
plot(
x,
prompt = FALSE,
na.rm = FALSE,
typographic = TRUE,
base_family = NULL,
...
)Arguments
- x
an object of class "compare_category", usually, a result of a call to compare_category().
- prompt
logical. The default value is FALSE. If there are multiple visualizations to be output, if this argument value is TRUE, a prompt is output each time.
- na.rm
logical. Specifies whether to include NA when plotting mosaics plot. The default is FALSE, so plot NA.
- typographic
logical. Whether to apply focuses on typographic elements to ggplot2 visualization. The default is TRUE. if TRUE provides a base theme that focuses on typographic elements.
- base_family
character. The name of the base font family to use for the visualization. If not specified, the font defined in dlookr is applied. (See details)
- ...
arguments to be passed to methods, such as graphical parameters (see par). However, it only support las parameter. las is numeric in 0, 1; the style of axis labels.
0 : always parallel to the axis [default],
1 : always horizontal to the axis,
Value
NULL. This function just draws a plot.
Details
The base_family is selected from "Roboto Condensed", "Liberation Sans Narrow", "NanumSquare", "Noto Sans Korean". If you want to use a different font, use it after loading the Google font with import_google_font().
Examples
# Generate data for the example
heartfailure2 <- heartfailure[, c("hblood_pressure", "smoking", "death_event")]
heartfailure2[sample(seq(NROW(heartfailure2)), 5), "smoking"] <- NA
# Compare the all categorical variables
all_var <- compare_category(heartfailure2)
# plot all pair of variables
plot(all_var)
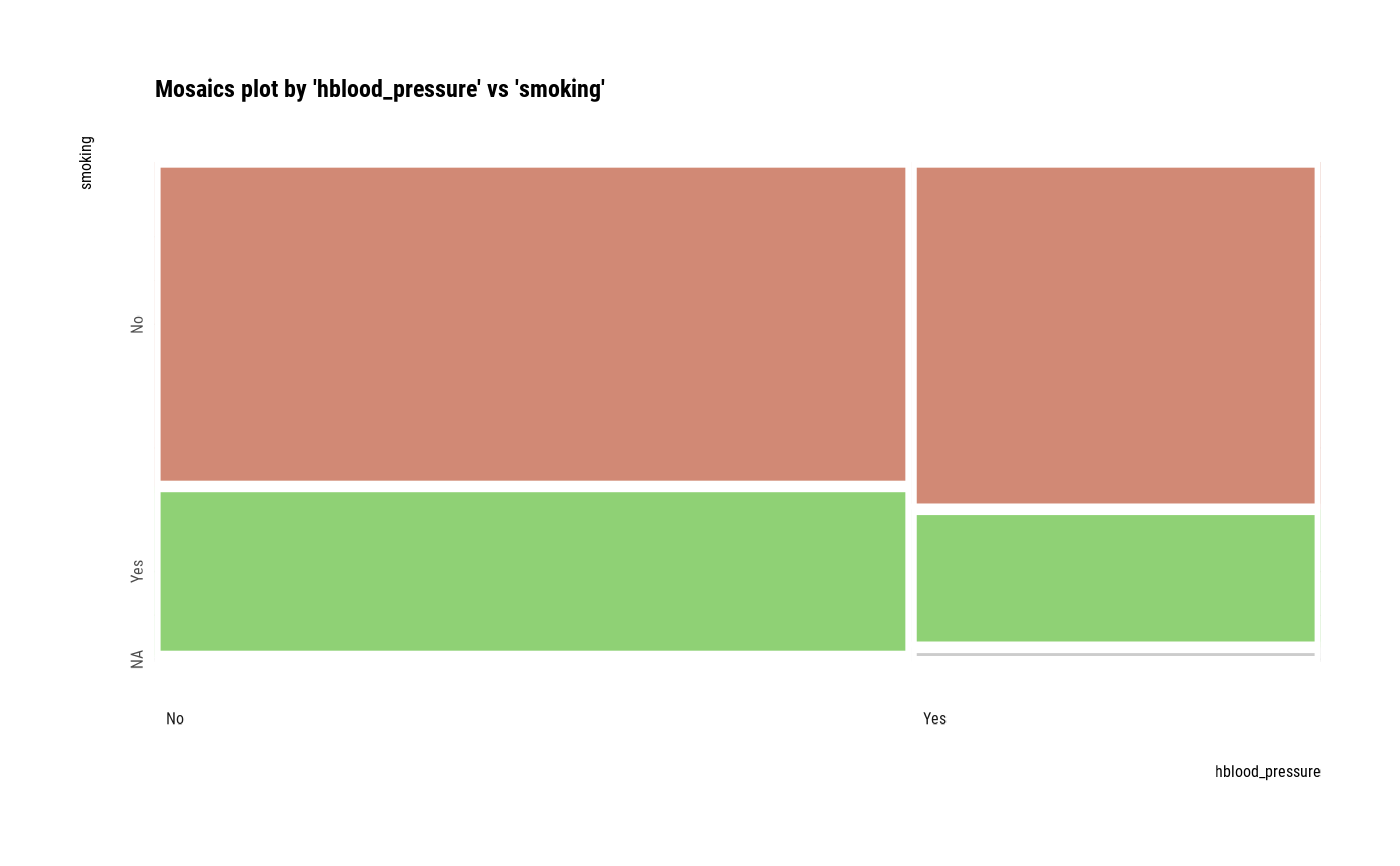
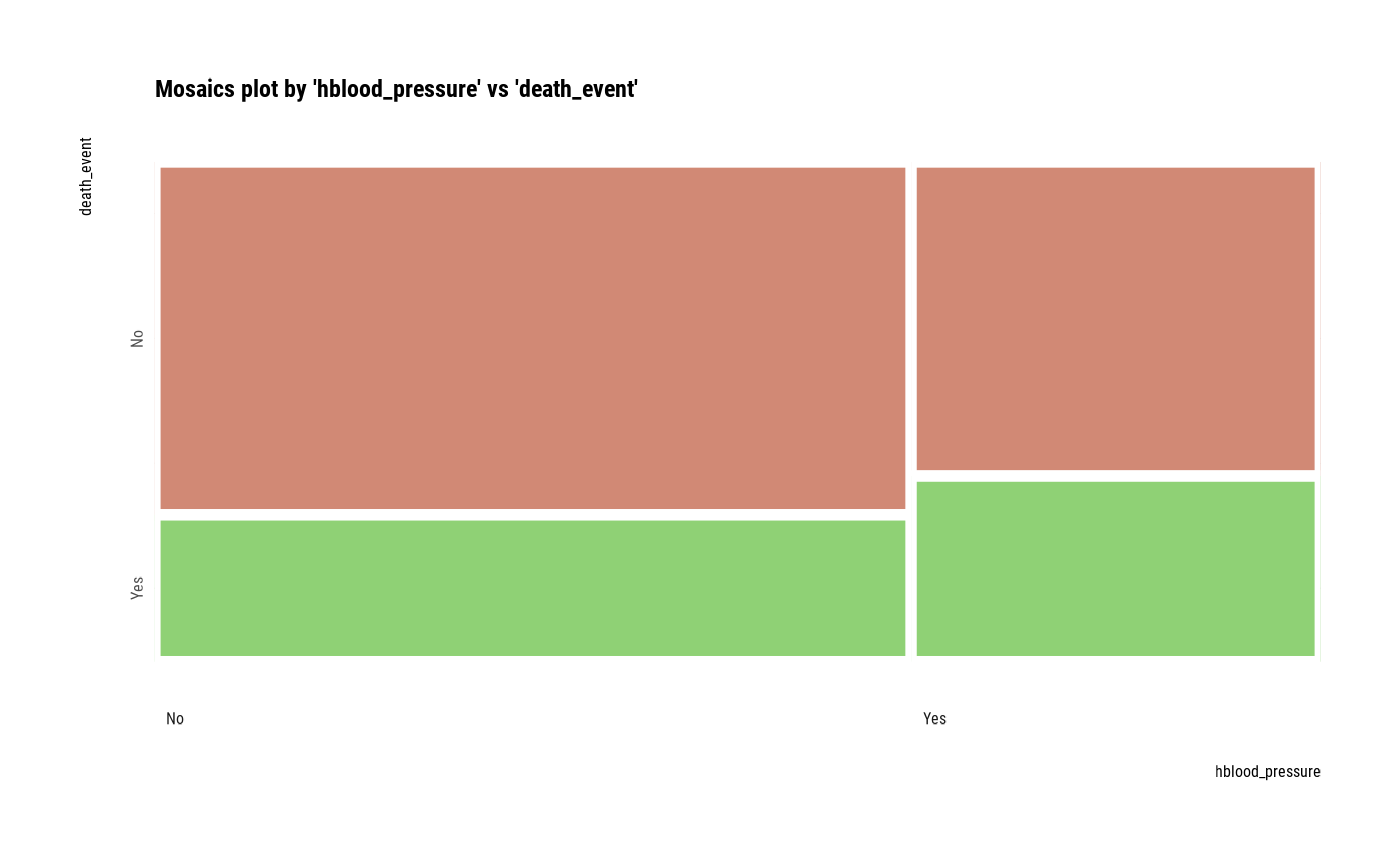
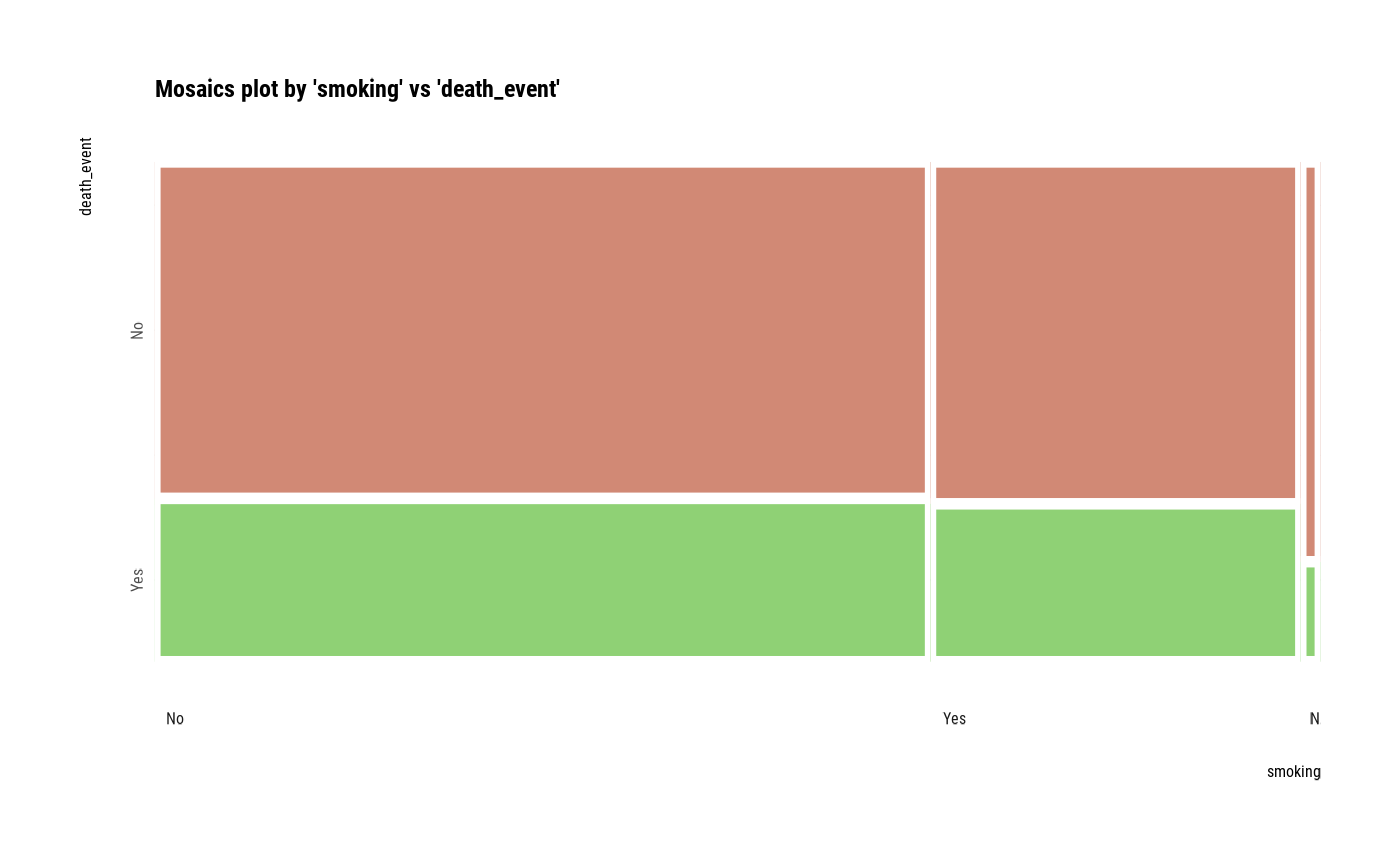 # Compare the two categorical variables
two_var <- compare_category(heartfailure2, smoking, death_event)
# plot a pair of variables
plot(two_var)
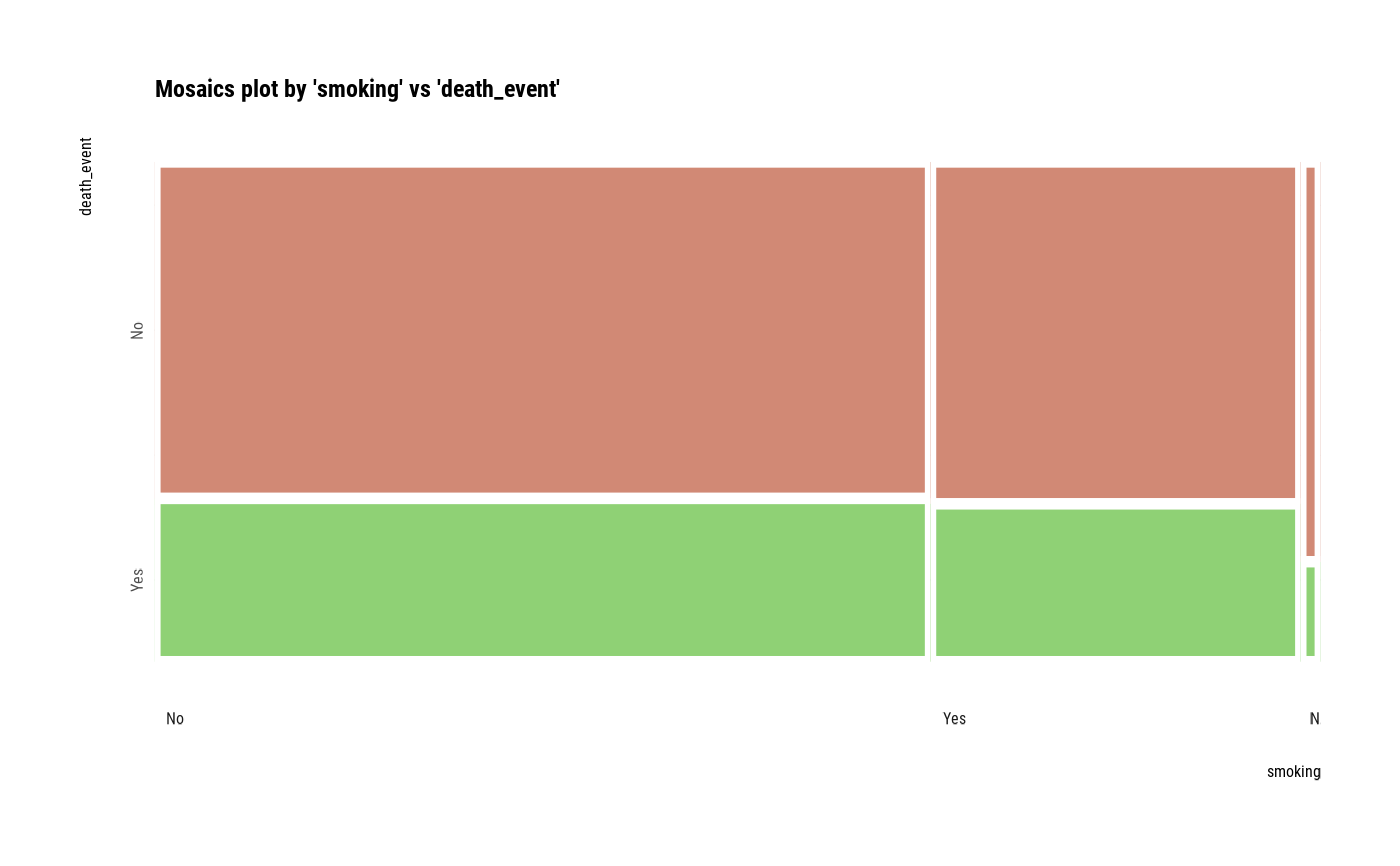
# Compare the two categorical variables
two_var <- compare_category(heartfailure2, smoking, death_event)
# plot a pair of variables
plot(two_var)
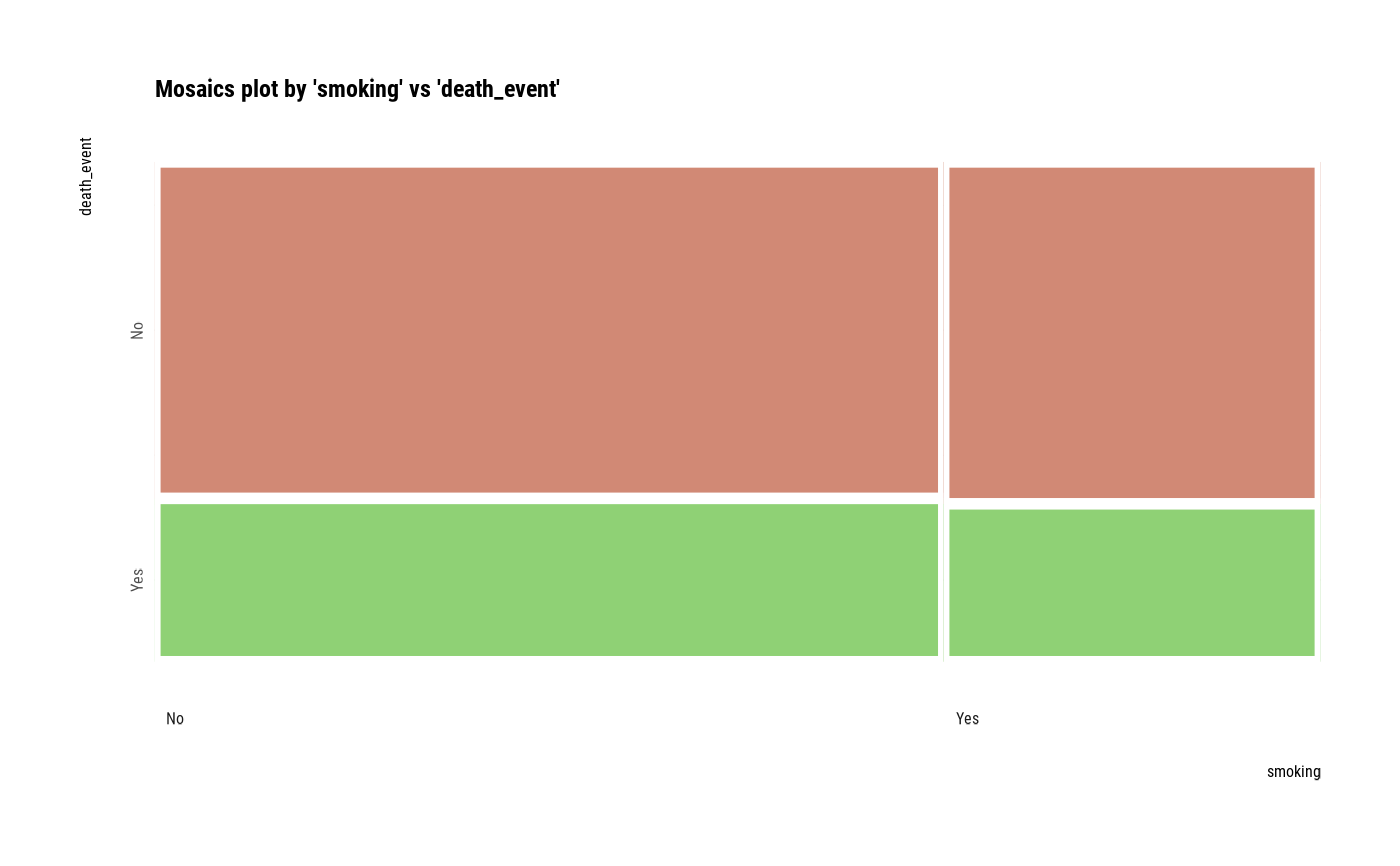
 # plot a pair of variables without NA
plot(two_var, na.rm = TRUE)
# plot a pair of variables without NA
plot(two_var, na.rm = TRUE)
 # plot a pair of variables not focuses on typographic elements
plot(two_var, typographic = FALSE)
# plot a pair of variables not focuses on typographic elements
plot(two_var, typographic = FALSE)